Systems neuroscience with Python: peering into the "black box"
Narendra Mukherjee
Brandeis University
About me
- 5th year PhD student in Neuroscience at Brandeis University
- Work with Prof. Donald Katz
- Study the processing of tastes in the rat brain
- Love Python (and R) and open source (and the bike!)
- More at: narendramukherjee.github.io
Systems neuroscience
Study of neural circuits and the brain 'systems' they form in the context of animal behavior.
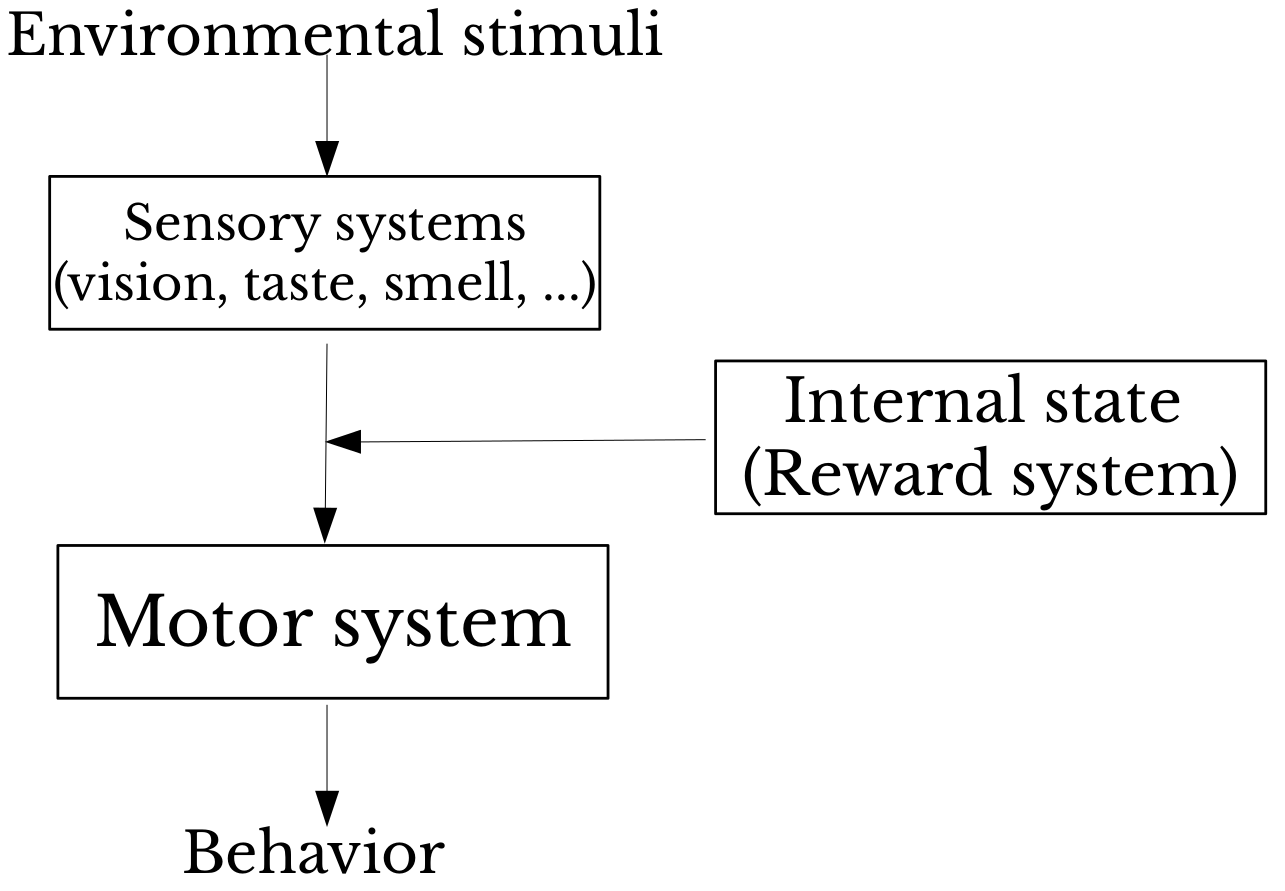
In-vivo electrophysiology (Ephys) is a crucial technique: recording the activity of neurons in awake, behaving animals.
The black box of Ephys
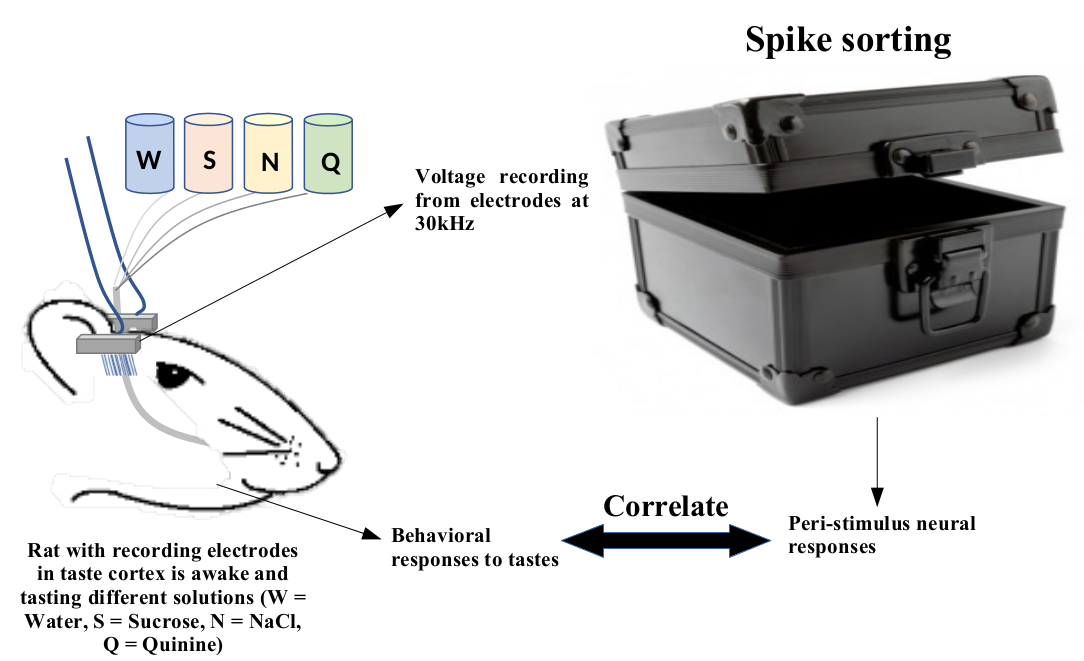
Peeking into the black box
Big data challenge!
- Voltage recordings from 64 electrodes at 30 kS/s (stored as 16 bit integers).
- (30000 samples) x (64 electrodes) x (2 bytes) ~ 3.8MB/s. About 26GB for a 2 hour recording.
- Electrode numbers/recording durations can potentially increase in the future.
Superpowers needed!
- Data format that can
- Allow fast access to small chunks of data and support larger-than-memory workloads.
- Structure data/metadata at different levels.
- Parallelization tool to break up the processing of each electrode's data into a separate process.
- GUI for users to provide inputs during spike sorting.
- Scientific computing stack for digital signal processing and unsupervised clustering.
- Diverse (and fast!) plotting framework.

Hierarchical Data Format (HDF5)
- Numerical big data.
- Data arranged in a hierarchy of nodes.
- Filesystem inside a file.
- Fast access to data with minimal I/O overhead.

Creating a HDF5 file
import tables # Create hdf5 file hf5 = tables.open_file(hdf5_name[-1]+'.h5', 'w', title = hdf5_name[-1]) # Node for raw electrode data hf5.create_group('/', 'raw') # Node for digital inputs hf5.create_group('/', 'digital_in') # Node for digital outputs hf5.create_group('/', 'digital_out') hf5.close() # Create an array with data hf5.create_array('/raw', 'electrode1', electrode1_data)
Pytables loves Numpy
The data is extracted as numpy arrays following the hierarchy of the nodes in the HDF5 file
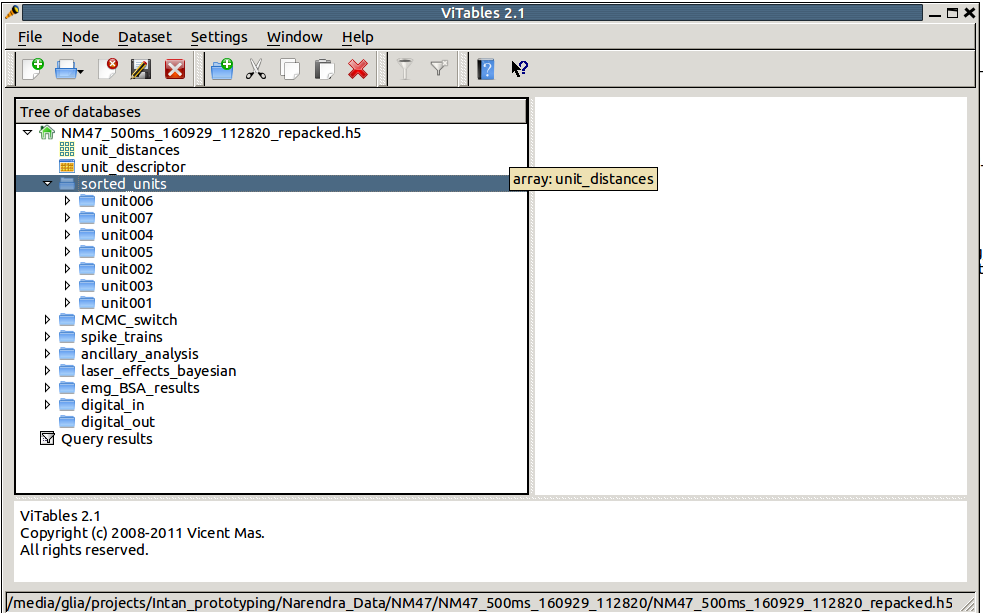
# Extract the spike waveforms of neuron 1 data = hf5.root.sorted_units.unit001.waveforms[:]
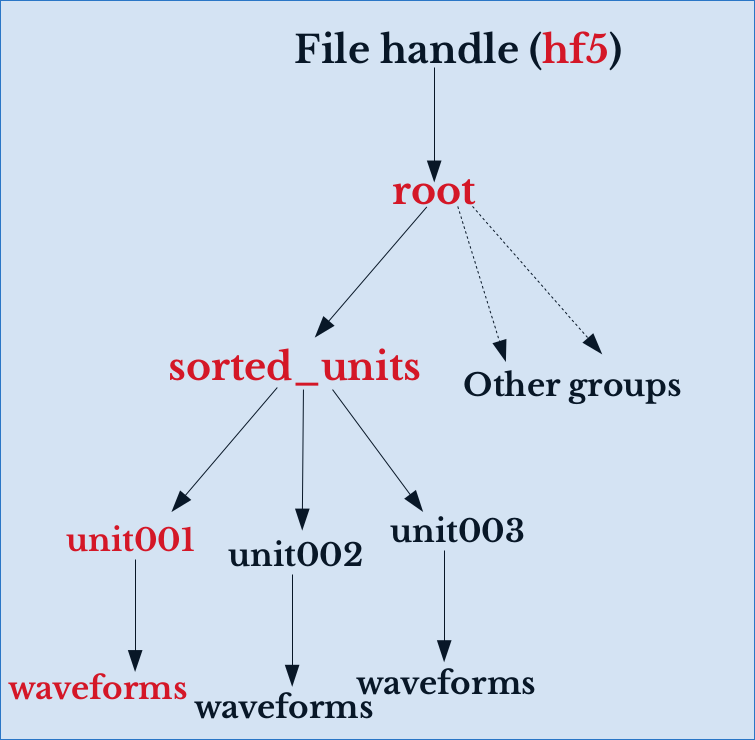
Meta-data resides in the same file
# Define a UnitDescriptor class to be used # to add things (anything!) about the sorted # units to a pytables table class UnitDescriptor(tables.IsDescription): electrode_number = tables.Int32Col() # Is this a single neuron? single_unit = tables.Int32Col() # 2 types of single neurons regular_spiking = tables.Int32Col() fast_spiking = tables.Int32Col()
More utilities with Pytables
- Compression tool: ptrepack
- Viewer tool: vitables
GNU Parallel for parallelizing processing
- Great parallelization tool for .nix systems.
- Allows processes to be released dependent on CPU workload, available RAM, etc.
- Can resume failed processes from a log file.
- Very similar feel to HPC task managers (like qsub).
GUI for user inputs: easygui
import easygui # Get name of directory with the data files dir_name = easygui.diropenbox() # Get the type of data files (.rhd or .dat) file_type = easygui.multchoicebox(msg = 'File type', choices = ('.dat', '.rhd'))
Data analysis stack

- Scipy for digital signal processing.
- Numpy for data manipulation.
- Scikit-learn for unsupervised clustering.
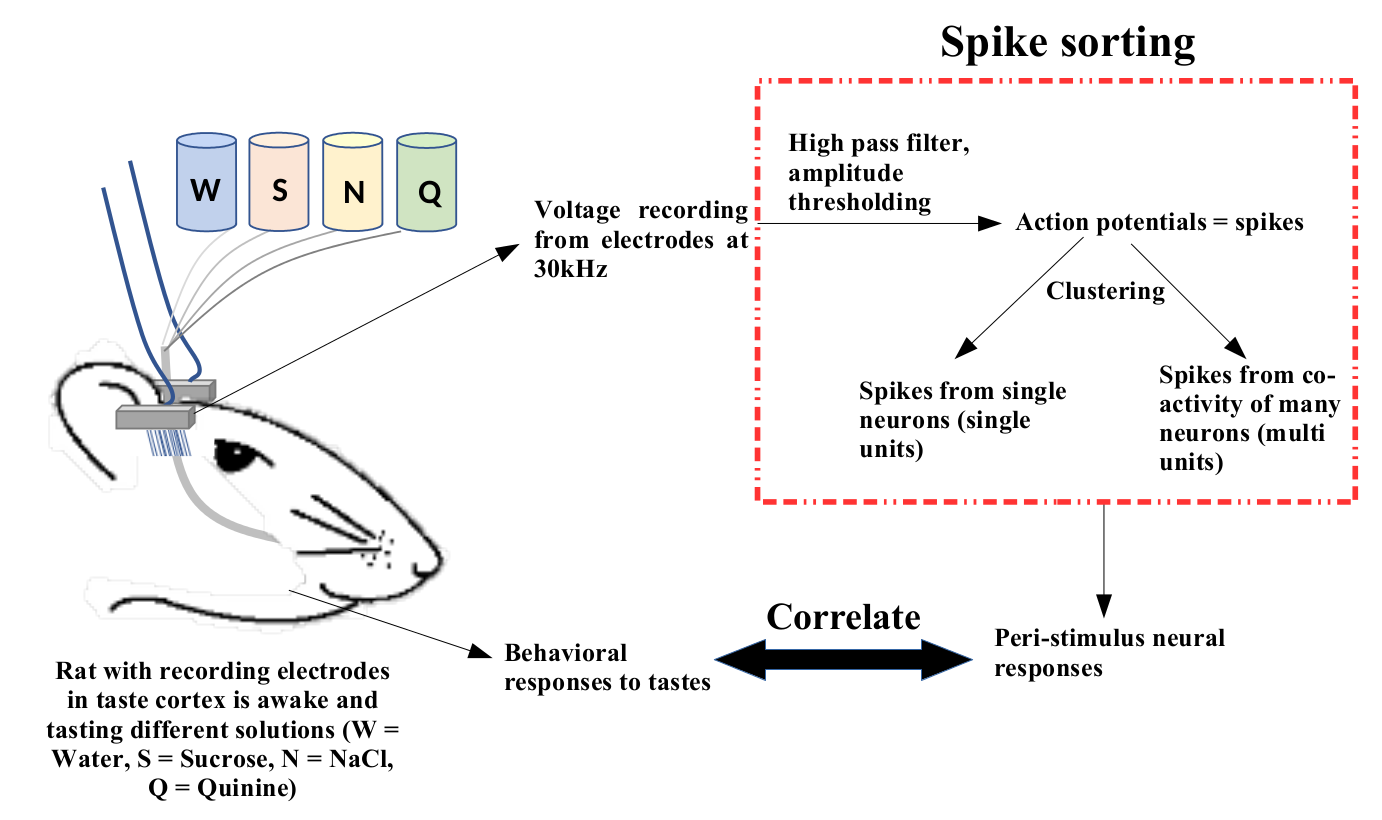
Spike sorting "transparent box"
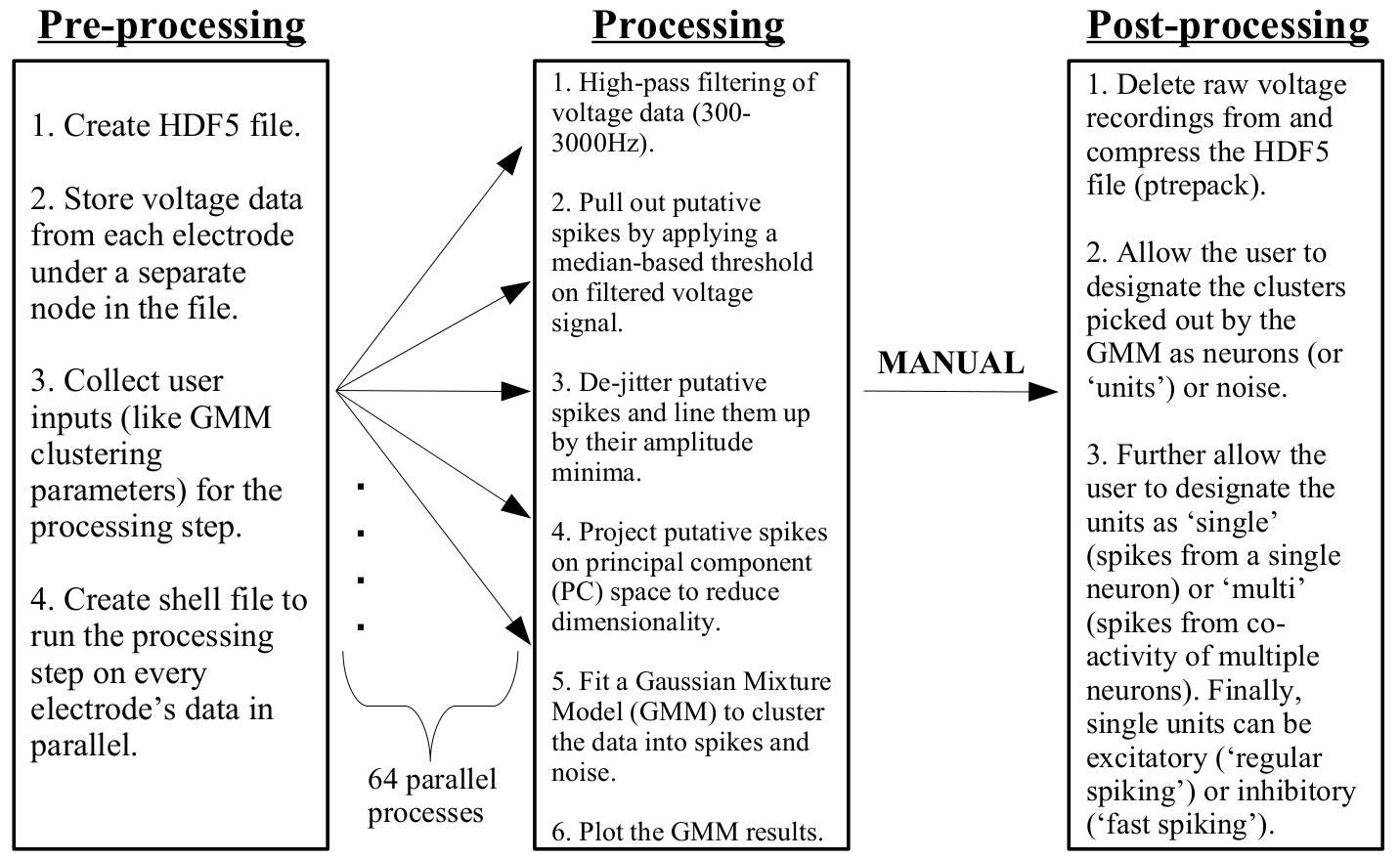
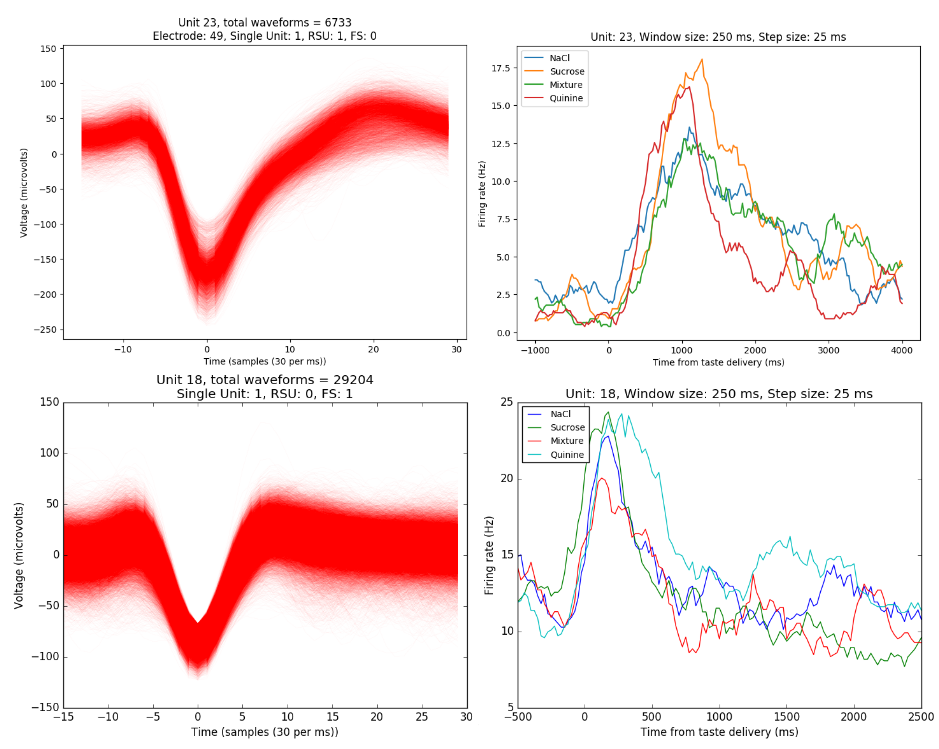
Results: spikes from single neurons
More details
Paper in Proceedings of Scipy 2017
Katz lab website
Github: narendramukherjee/blech_clust
Email: narendra(at)brandeis(dot)edu
All thanks to...


